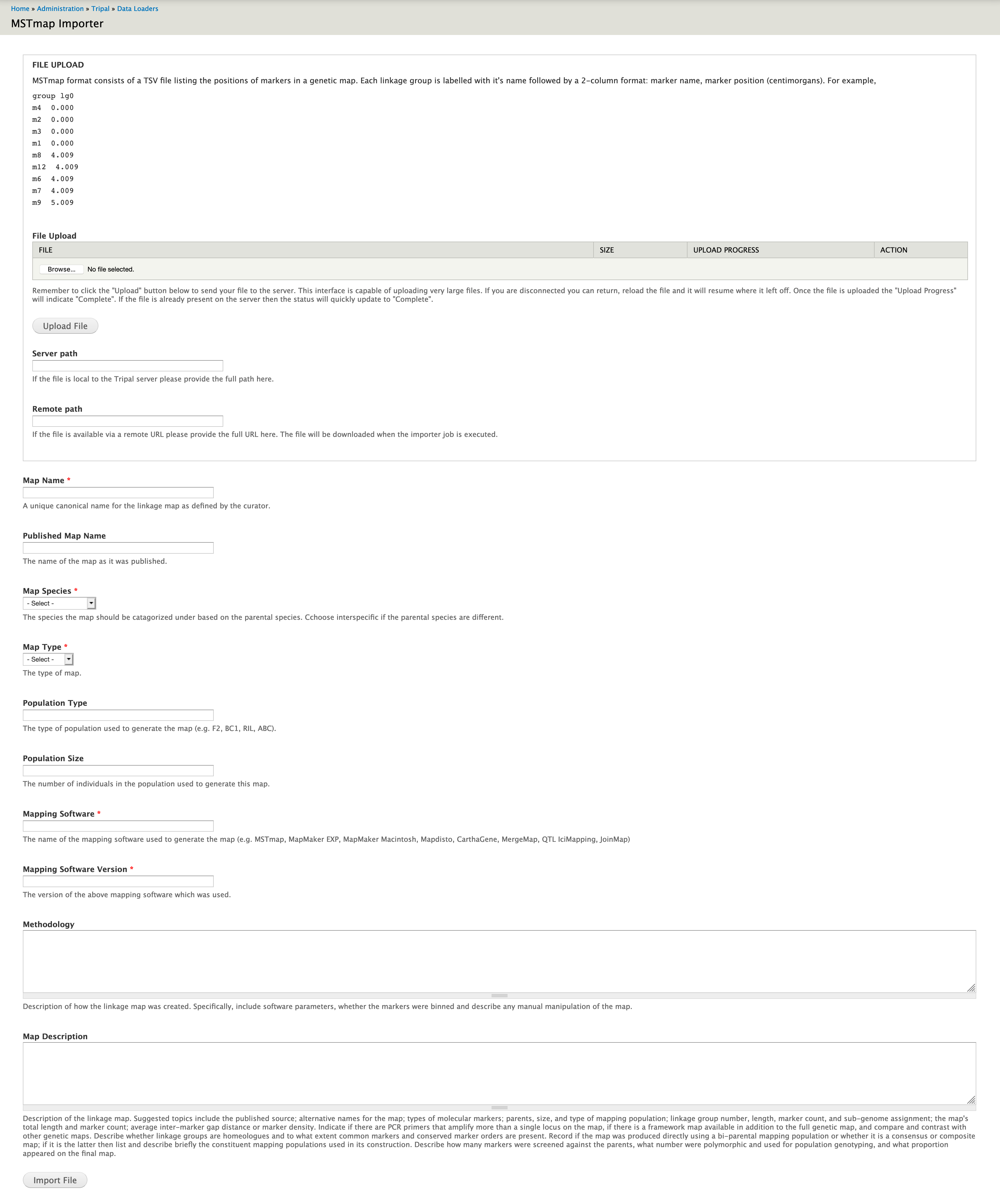
MSTmap Importer¶
Format¶
This importer will load genetic maps which follow the same format output by the MSTmap software. This format consists of a TSV file listing the positions of markers in a genetic map. Each linkage group is labelled with it’s name followed by a 2-column format: marker name, marker position (centimorgans). For example,
group lg0
m4 0.000
m2 0.000
m3 0.000
m1 0.000
m8 4.009
m12 4.009
m6 4.009
m7 4.009
m9 5.009
The importer also stores metadata about the map through a well described form:
Validation¶
There is currently no validation of the file on import. However, the metadata in the form is minimally validated (e.g. the organism must already exist in order to be chosen).
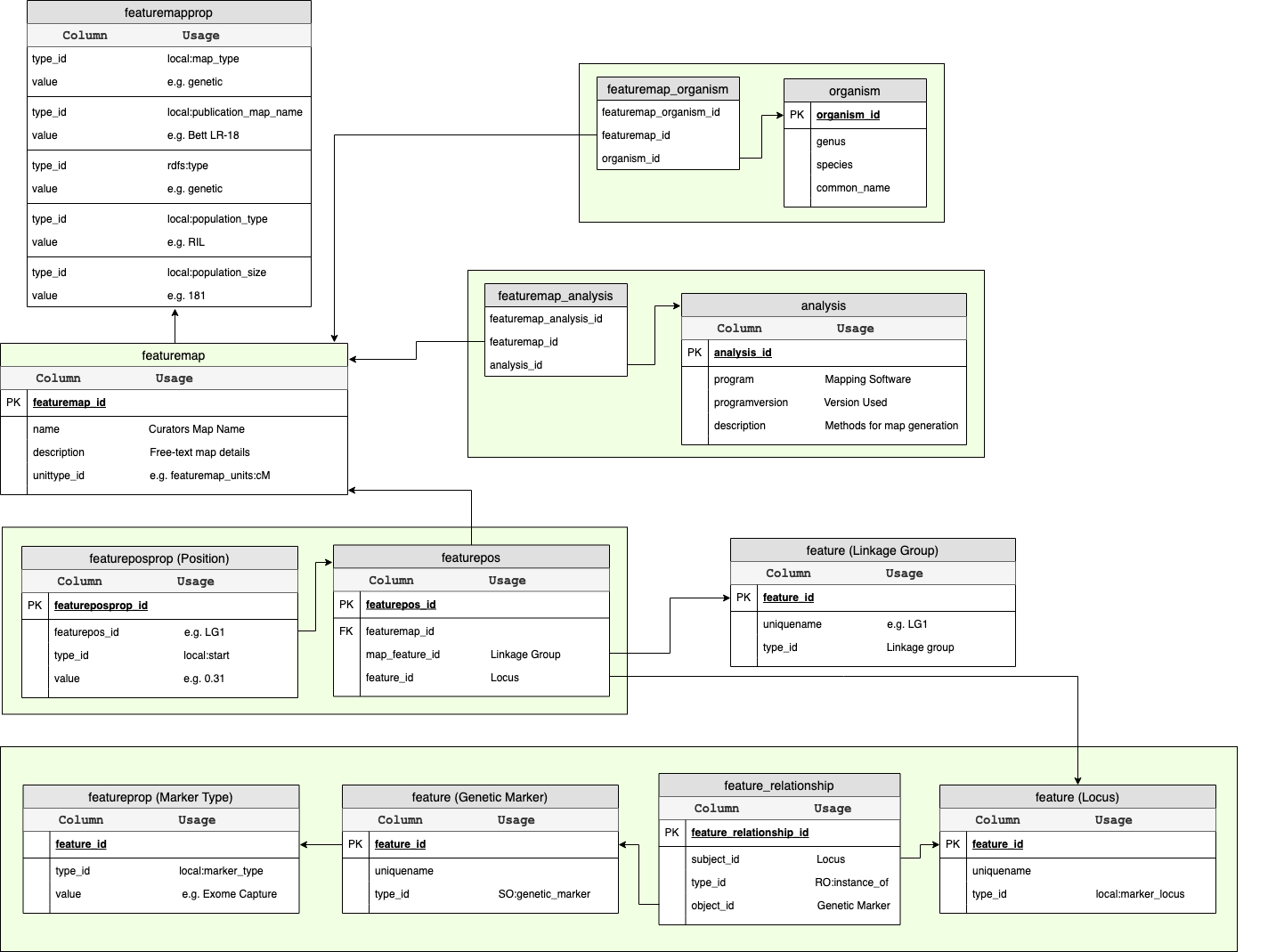
Data Storage¶
Metadata about the map including it’s name, description, population type and size are stored in the featuremap and featuremapprop tables of chado; whereas, metadata describing the methodology is stored in the analysis table. Each position in the map is a combination of the linkage group, locus and genetic marker (features) with the position itself stored in the featurepos table. This is more clearly explained via the ER diagram below.